Semi-prestained Protein Marker
|
Semi-prestained Protein Marker |
 Technical Brochure Technical Brochure |
| Description: The semiprestained Protein Marker is a mixture of 8 highly purified unstained proteins and 3 prestained proteins (16, 34, 85 kDa) that resolve into 11 identifiable bands from 14.4-85 kDa when analysed by SDS-PAGE and stained with Coomassie Brilliant Blue R-250, or the protein stain of choice. This marker is intended for use as a precise size standard when performing SDS-PAGE in order to calculate the molecular weight of a protein of your interest and as marker for western blot experiments.
Preparation: Gently mix the semiprestained Protein Marker by vortexing or pipetting up and down several times. Pipette the desired amount (5-7μl/lane) to a separate tube. Incubate at 95-100oC for 5 minutes, spin down and load.
Storage buffer: 62,5mM Tris-HCl (pH=6,8), 2% SDS, 1mM EDTA, 30% Glycerol, 0,01% Bromophenol blue, 1mM NaN3, 50mM DTT. Store at -20oC.
Note:
. |
|
T7 RNA polymerase
|
T7 RNA polymerase (concentration 50U/μl) |
 technical brochure technical brochure |
|||||||||||||||||||
|
Source: Purified from an E. coli strain carrying a plasmid with the cDNA o RNA polymerase of T7 phage. Description: MINOTECH T7 RNA polymerase is an efficient RNA polymerase able to transcribe DNA sequences under the T7 phage promoter up to (5.5 kbs). Unit definition: One unit is defined as the amount of enzyme that will incorporate 1 nmol ATP into acid-insoluble material in a total reaction volume of 50 μl in 1 hour at 37°C. Quality Control Assays:
Guaranteed stability: T7 RNA polymerase is guaranteed to maintain stability until expiration date.
|
*PCR products/annealed oligos or plasmid DNA containing T7 promoter
Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, p. 194-197. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y |
|||||||||||||||||||
MINOTECH RT
|
MINOTECH RT (concentration 200U/μl) |
 technical brochure technical brochure |
||||||||||||||||||||||||||||||||||||||||||||||||
|
Source: Purified from an E. coli strain carrying a plasmid with M-MuLV reverse transcriptase gene. Description: High purity reverse transcriptase suitable for first strand cDNA synthesis. Unit definition: One unit is defined as the amount of enzyme required to incorporate 1 nmol of dTTP into acid-insoluble material in a total reaction volume of 50 μl in 10 minutes at 37°C using poly(rA)•oligo(dT)18 as template. Quality Control Assays:
Guaranteed stability: MINOTECH RT is guaranteed to maintain stability until expiration date. Recommended First-Strand cDNA Synthesis mixture:
* 400 U of MINOTECH RT can be added to increase yield (for the generation of cDNA >5kb).
Optional (recommended for PCR targets >1kb). Remove RNA complementary to the cDNA, by adding 2 units of E. coli RNase H and incubate at 37°C for 20 minutes. |
|
||||||||||||||||||||||||||||||||||||||||||||||||
XbaI
| XbaI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
XbaI is a restriction enzyme purified from Xanthomonas badrii.
Unit substrate: Lambda DNA (dam-/HindIII digest).
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 200 units of XbaI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNAdam-/HindIII digest at 37oC. After 100-fold overdigestion with XbaI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol , 500 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Blocked by overlapping dcm methylation: Not sensitive CpG methylation: Not sensitive |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
TaqI
| TaqI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
TaqI is a restriction enzyme purified from Thermus aquaticus YT I.
Unit substrate: Lambda DNA (dam-).
Unit calculation assay conditions: 100 mM KCl, 20 mM Tris-HCl (pH 8.5 @ 25°C), 3 mM MgCl2, 0.04% Triton X-100, 100 μg/ml BSA. Incubate at 65oC.
Absence of contaminants: 100 units of TaqI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA (dam-) at 65oC. After 50-fold overdigestion with TaqI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 300 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 500 μg/ml BSA and 50% glycerol. Store at -20oC
Heat inactivation: 80oC for 20 minutes.
Methylation Sensitivity: dam methylation: Blocked by overlapping dcm methylation: Not sensitive CpG methylation: Not sensitive Note: Incubation without BSA results in 50% activity. |
Reagents supplied: 10x UTaqI buffer
|
||||||||||||||||||||||||||||||||||||||
StyI
| StyI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
StyI is a restriction enzyme purified from E.coli WA921/pST27 hsd+.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 50 units of StyI do no produce any unspecific clevage products after 16 hrs incubation with 1 μg of λ DNA at 37°C. After 50-fold overdigestion with StyI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration >5%, or pH>8.0 may result in star activity |
Reagents supplied: 10x H buffer
|
||||||||||||||||||||||||||||||||||||||
SstI (SacI isoschizomer)
|
SstI (SacI isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SstI is a restriction enzyme purified from Streptomyces stanford.
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 100 units of SstI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA/HindIII digest at 37oC. After 50-fold overdigestion with SstI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive |
Reagents supplied: 10x L and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
CspA I (Age I isoschizomer)
| CspA I (Age I isoschizomer) |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
CspAI is a restriction enzyme purified from Corynebacterium species.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 10 mM Bis Tris Propane-HCl (pH 7.0 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 50 units of CspA I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 10-fold overdigestion with CspA I, greater than 90% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive |
Reagents supplied: 10x UCspAI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
BstE II
| BstE II |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BstEII is a restriction enzyme purified from Bacillus stearothermophilus.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM KCl, 10 mM Tris-HCl (pH 7.4 @ 25°C), 5 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100, 100 μg/ml BSA. Incubate at 60oC.
Absence of contaminants: 150 units of BstEII do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 60oC. After 100-fold overdigestion with BstEII, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Note: BstE II exhibits 10-15% activity at 37°C. |
Reagents supplied: 10x UBstEII and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
SspI
| SspI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SspI is a restriction enzyme purified from Sphaerotilus species.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 30 units of SspI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 37°C. After 10-fold overdigestion with SspI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration >5%, or pH>8.0 may result in star activity. |
Reagents supplied: 10x H and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
BssAI (Cfr10 I isoschizomer)
| BssAI (Cfr10 I isoschizomer) |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BssAI is a restriction enzyme purified from Bacillus species.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM KCl, 20 mM Tris-HCl (pH 8.5 @ 25oC), 3 mM MgCl2, 0.04% Triton X-100, 100 μg/ml bovine serum albumin and DNA. Incubate at 65oC.
Absence of contaminants: 50 units of BssA I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 65oC. After 30-fold overdigestion with BssA I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Reference: Rina, M., Stratidakis, I. and Bouriotis, V. (1990). Nucleic Acids Res. 18, 6161. |
Reagents supplied: 10x UBssAI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
SseBI (Stu I isoschizomer)
|
SseBI (Stu I isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
SseBI is a restriction enzyme purified from Streptomyces species.
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA and DNA. Incubate at 37oC.
Absence of contaminants: 150 units of SseBI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA/Hind III digest at 37oC. After 50-fold overdigestion with SseBI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Blocked by overlapping CpG methylation: Not sensitive Reference: Rina, M., Tzanodaskalaki, M., Karagouni, A., Pagomenou, M. and Bouriotis, V. (1992) Nucleic Acids Res. 20, 1808. |
Reagents supplied: 10x H and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
SphI
| SphI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SphI is a restriction enzyme purified from Streptomyces phaeochromogenes.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 50 units of SphI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 37oC. After 10-fold overdigestion with SphI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 400 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
BshF I (Hae III isoschizomer)
|
BshF I (Hae III isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BshFI is a restriction enzyme purified from Bacillus sphaericus.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM potassium acetate, 20 mM Tris-acetate (pH 7.9 @ 25oC), 10 mM magnesium acetate, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 200 units of BshF I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 50-fold overdigestion with BshF I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 80oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Reference: Vlatakis, G., Clark, D. and Bouriotis, V. (1989). Nucleic Acis Res. 17, 8882 |
Reagents supplied: 10x A and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
BseC I (Cla I isoschizomer)
| BseC I (Cla I isoschizomer) |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BseCI is a restriction enzyme purified from Bacillus stearothermophilus.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 55oC.
Absence of contaminants: 150 units of BseCI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 55oC. After 100-fold overdigestion with BseCI greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 0.1 mM EDTA, 1 mM dithiothreitol, 0.15% Triton X-100, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Blocked by overlapping dcm methylation: Not sensitive CpG methylation: Blocked
Reference: Rina, M., Dialektakis, D., Clark, D., Pagomenou, M. and Bouriotis, V. (1992). Nucleic Acids Res. 20 |
Reagents supplied: 10x H and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
SmaI
| Sma I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SmaI is a restriction enzyme purified from Serratia marcescens (ATCC 49779).
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 50 mM potassium acetate, 20 mM Tris-acetate (pH 7.9 @ 25°C), 10 mM magnesium acetate, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 25oC.
Absence of contaminants: 150 units of SmaI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA (HindIII digest) at 25oC. After 50-fold overdigestion with SmaI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked |
Reagents supplied: 10x A and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Sla I (Xho I isoschizomer)
|
Sla I (Xho I isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SlaI is a restriction enzyme purified from Stremptomyces lavendulae.
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 150 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 400 units of Sla I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA (Hind III digest) at 37oC. After 100-fold overdigestion with SlaI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Impaired |
Reagents supplied: 10x SH and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
SgrBI (SacII isoschizomer)
|
SgrBI (SacII isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SgrBI is a restriction enzyme purified from Streptomyces griseus.
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.9@ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 400 units of SgrB I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA (HindIII digest) at 37°C. After 100-fold overdigestion with SgrBI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked
Note: Particular sites in λ and φΧ174 DNAs are difficult to cleave with SgrB I, as well as with its prototype Sac II. Reference: Rina, M., Pagomenou, M. and V, Bouriotis (1991) Nucleic Acids Res. 19, 6342. |
Reagents supplied: 10x USgrBI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Sfi I
| Sfi I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SfiI is a restriction enzyme purified from Streptomyces fimbriatus (ATCC 15051).
Unit substrate: Adenovirus-2 DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 50°C.
Absence of contaminants: 100 units of Sfi I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Adeno-2 DNA at 50oC. After 50-fold overdigestion with Sfi I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 300 mM NaCl, 5 mM KPO4 (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 0.15% Triton X-100, 500 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Impaired by overlapping CpG methylation: Blocked by some combinations of overlapping |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Sca I
| Sca I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| ScaI is a restriction enzyme purified from Streptomyces caespitosus.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM KCl, 10 mM Bis-Tris-Propane (pH 7.0 @ 25°C), 10 mM MgCl2, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 600 units of Sca I incubated for 16 hours at 37ºC with 1 μg of λ DNA resulted in a DNA pattern free of detectable nuclease degradation as determined by agarose gel electrophoresis. After 100-fold overdigestion with Sca I, greater than 90% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 500 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 80oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Star activity: Large excess of the enzyme may results in the appearance of star activity. |
Reagents supplied: 10x UScaI buffer
|
||||||||||||||||||||||||||||||||||||||
Pst I
| Pst I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| PstI is a restriction enzyme purified from a recombinant E.coli strain.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.4 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 200 units of Pst I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 100-fold overdigestion with Pst I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 200 mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin, 0.15% Triton X-100 and 50% glycerol. Store at -20oC.
Heat inactivation: 80oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive Star activity: Conditions of high enzyme concentration or glycerol concentration>12% may result in star activity. |
Reagents supplied: 10x UPstI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Sau3AI
| Sau3AI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| Sau3AI is a restriction enzyme purified from Streptomyces species.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1mM dithiothreitol, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 50 units of Sau3A I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 37oC. After 50-fold overdigestion with Sau3A I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by overlapping |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
PspP I (Sau96 I isoschizomer)
| PspP I (Sau96 I isoschizomer) |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| PspPI is a restriction enzyme purified from Psychrobacter immobilis TA137.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 25oC.
Absence of contaminants: 80 units of PspP I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 25oC. After 50-fold overdigestion with PspP I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 55oC for 15 minutes..
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Blocked by overlapping CpG methylation: Blocked by overlapping
Note: Incubation at 37oC results in 60% activity.
Reference: Rina, M., Caufrier, F., Mavromatis, K., Markaki, M., Kokkinidis, M. and Bouriotis, V. (1997) Gene, 197, 353-360. |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Sal I
| Sal I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SalI is a restriction enzyme purified from Streptomyces albus G.
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 150 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 400 units of Sal I incubated for 16 hours at 37ºC with 1 μg of λ DNA (HindIII digest) resulted in a DNA pattern free of detectable nuclease degradation as determined by agarose gel electrophoresis. After 50-fold overdigestion with Sal I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 300 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked
Star activity: Large excess of the enzyme results in the appearance of star activity. |
Reagents supplied: 10x SH buffer
|
||||||||||||||||||||||||||||||||||||||
Nru I
| Nru I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| NruI is a restriction enzyme purified from Nocardia rubra.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM KCl, 50 mM Tris-HCl (pH 8.0 @ 25oC), 10 mM MgCl2, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 80 units of Nru I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 10-fold overdigestion with Nru I, less than 20% of the DNA fragments can be ligated.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Blocked by overlapping dcm methylation: Not sensitive CpG methylation: Blocked
Star activity: Large excess of the enzyme results in the appearance of star activity. |
Reagents supplied: 10x UNruI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Not I
| Not I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
NotI is a restriction enzyme purified from Nocardia otitidis-caviarum.
Unit substrate: Adenovirus-2 DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25°C), 5 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 80 units of Not I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Adeno-2 DNA at 37°C. After 30-fold overdigestion with Not I, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 500 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1mM dithiothreitol, 0.1% Triton X-100, 500 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked Note: Supercoiled plasmids may require up to 5-fold more Not I for complete digestion than linear DNAs. |
Reagents supplied: 10x UNotI buffer
|
||||||||||||||||||||||||||||||||||||||
RsaI
| RsaI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| RsaI is a restriction enzyme purified from Rhodopseudomonas sphaeroides.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 400 units of Rsa I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 37oC. After 10-fold overdigestion with Rsa I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by some combinations of overlapping
Note: Cleaves single-stranded DNA slowly. |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Nhe I
| Nhe I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| NheI is a restriction enzyme purified from Neisseria mucosa heildelbergensis (ATCC 25999).
Unit substrate: Lambda DNA (HindIII digest).
Unit calculation assay conditions: 50 mM potassium acetate, 20 mM Tris-acetate (pH 7.9 @ 25°C), 10 mM magnesium acetate, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 80 units of NheI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA/Hind III digest at 37oC. After 100-fold overdigestion with NheI, greater than 98% of the DNA fragments can be ligated and recut.
Storage buffer: 200 mM NaCl, 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 1 mM dithiothreitol, 0.15% Triton X-100, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65°C for 20 minutes..
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by some combinations of overlapping
Note: Activity inhibited by salt concentrations >100mM. Cleaves to leave a 5’ CTAG extension which can be efficiently ligated to DNA fragments generated by AvrII, SpeI, or XbaI. Star activity: Low salt, high glycerol (>5%) concentrations, pH >8.0 or large excess of the enzyme may result in star activity. |
Reagents supplied: 10x A and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Nco I
| Nco I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| NcoI is a restriction enzyme purified from Nocardia corallina.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM DTT, 0.02% Triton X-100, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 100 units of Nco I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 50-fold overdigestion with Nco I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive |
Reagents supplied: 10x UNcoI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Pvu II
| PvuII |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| PvuII is a restriction enzyme purified from a recombinant E.coli strain.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 100 units of Pvu II do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 10-fold overdigestion with Pvu II greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration >5% or pH >8.0 may result in star activity. |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Nae I
| Nae I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| NaeI is a restriction enzyme purified from Streptomyces species.
Unit substrate: pBR322 DNA.
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 50 units of Nae I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of pBR322 at 37oC. After 10-fold overdigestion with Nae I, greater than 80% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked Note: Nae I exhibits site preferences. pBR322 contains four Nae I recognition sequences. Two of these sites are readily cleaved, one is cleaved moderately slowly, and the fourth is cleaved 50-fold more slowly. |
Reagents supplied: 10x L and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
BseB I (BstN I isoschizomer)
| BseB I (BstN I isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BseBI is a restriction enzyme purified from Bacillus stearothermophilus.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH @ 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 60°C.
Absence of contaminants: 500 units of BseBI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 60oC. After ten-fold overdigestion with BseBI, less than 50% of the DNA fragments can be ligated.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Note: BseBI-cut DNA is difficult to ligate with T4 DNA Ligase. Ligation is enhanced in the presence of 15% PEG4000. |
Reagents supplied: 10x M buffer
|
||||||||||||||||||||||||||||||||||||||
BseA I (BspMII isoschizomer)
|
BseA I (BspMII isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BseAI is a restriction enzyme purified from Bacillus stearothermophilus.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 10 mM Tris-HCl (pH 8.0 @ 25°C), 5 mM MgCl2, 1 mM dithiothreitol, 0.02% Triton X-100, 100 μg/ml BSA. Incubate at 55oC.
Absence of contaminants: 400 units of BseAI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 55oC. After 100-fold overdigestion with BseAI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 500 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Reference: Thanos, D., Scarpelis, G., Papamatheakis, J. and Bouriotis, V. (1989). Nucleic Acids Res. 17, 8881. |
Reagents supplied: 10x UBseAI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
MspC I (Afl II isoschizomer)
|
MspC I (Afl II isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
MspCI is a restriction enzyme purified from Micrococcus species.
Unit substrate: Lambda DNA (Hind III digest).
Unit calculation assay conditions: 150 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 80 units of MspCI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA/Hind III digest at 37oC. After 10-fold overdigestion with MspCI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive Reference: Rina, M., Tzanodaskalaki, M., Karagouni, A., Pagomenou, M. and Bouriotis, V. (1992). Nucleic Acids Res., 20, 1806. |
Reagents supplied: 10x SH and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Bgl II
| Bgl II |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BglII is a restriction enzyme purified from Bacillus globigii lacking BglI.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 150 units of BglII do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 37oC. After 50-fold overdigestion with BglII, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive |
Reagents supplied: 10x H and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Mbo I
| Mbo I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| MboI is a restriction enzyme purified from Moraxella bovis (ATCC 10900).
Unit substrate: Lambda DNA (dam-).
Unit calculation assay conditions: 100 mM KCl, 10 mM Tris-HCl (pH 8.0 @ 25°C), 10 mM MgCl2, 1mM dithiothreitol, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 20 units of Mbo I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA (dam–) at 37oC. After 10-fold overdigestion with Mbo I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes..
Methylation Sensitivity: dam methylation: Blocked dcm methylation: Not sensitive CpG methylation: Impaired by overlapping |
Reagents supplied: 10x UMboI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Kpn I
| Kpn I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| KpnI is a restriction enzyme purified from Klebsiella pneumonia OK8.
Unit substrate: Lambda DNA (EcoRI digest).
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.0 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 0.01% Triton X-100, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 30 units of Kpn I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of lambda DNA/EcoR I digest at 37oC. After 10-fold overdigestion with KpnI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration >5% or pH >8.0 may result in star activity. |
Reagents supplied: 10x UKpnI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Bgl I
| Bgl I |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BglI is a restriction enzyme purified from Bacillus globigii lacking BglII.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM KCl, 20 mM Tris-HCl (pH 8.5 @ 25°C), 10 mM MgCl2, 0.04% Triton X-100, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 100 units of BglI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA at 37oC. After 50-fold overdigestion with BglI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 300 mM NaCl, 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 1 mM dithiothreitol, 500 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by some combinations of overlapping |
Reagents supplied: 10x UBglI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
HpaI
| HpaI |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| HpaI is a restriction enzyme purified from a recombinant E.coli strain.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM potassium acetate, 20 mM Tris-acetate (pH 7.9 @ 25oC), 10 mM magnesium acetate, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 50 units of Hpa I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 10-fold overdigestion with Hpa I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 500 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by some combinations of overlapping Star activity: Conditions of high enzyme concentration or glycerol concen-tration >5%, may result in star activity. |
Reagents supplied: 10x A and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Hinf I
| Hinf I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
HinfI is a restriction enzyme purified from a recombinant E.coli strain.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 50 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 200 units of Hinf I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 100-fold overdigestion with Hinf I, greater than 90% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 80oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by some combinations of overlapping |
Reagents supplied: 10x H and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
Hind III
| Hind III |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| HindIII is a restriction enzyme purified from a recombinant E.coli strain.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 700 units of Hind III do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 100-fold overdigestion with Hind III, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 500 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Star activity: Star activity may be observed in the presence of Mn2+. |
Reagents supplied: 10x M and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
EcoR V
| EcoR V |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| EcoRV is a restriction enzyme purified from E. coli J62plg 74.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 100 units of EcoR V do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 20-fold overdigestion with EcoR V, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 80oC for 20 minutes..
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked by overlapping
Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration>5%, or pH>8.0 may result in star activity. |
Reagents supplied: 10x M and 10x K buffer |
||||||||||||||||||||||||||||||||||||||
EcoR I
| EcoR I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
EcoRI is a restriction enzyme purified from E. coli RY 13.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 50 mM NaCl, 100 mM Tris-HCl (pH 7.4 @ 25oC), 5 mM MgCl2, 0.025% Triton X-100, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 100 units of EcoR I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of Lambda DNA at 37oC. After 50-fold overdigestion with EcoR I greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 300 mM NaCl, 5 mM KPO4, (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 0.15% Triton X-100, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Impaired by overlapping Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration>5%, or pH>8.0 may result in star activity. |
Reagents supplied: 10x UEcoRI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
anti-GFP
|
anti-GFP |
|
|
Form: Serum (0.02% sodium azide) Source: α- GFP specific rabbit IgG Antigen: recombinant GFP Applications: Western blot analysis (detection by chemiluminescence): used titer 1:100,000.
Storage: Store at -20oC |
anti-RFP
|
anti-RFP |
|
|
Form: Serum (0.02% sodium azide) Source: α- RFP specific rabbit IgG Antigen: recombinant mRFP1 Applications: Western blot analysis (detection by chemiluminescence): used titer 1:100,000.
Storage: Store at -20oC |
BclI
| Bcl I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BclI is a restriction enzyme purified from Bacillus caldolyticus.
Unit substrate: Lambda DNA (dam-).
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 50oC.
Absence of contaminants: 100 units of BclI do no produce any unspecific clevage products after 16 hrs incubation with 1 μg of λ DNA (dam-) at 50°C. After 50-fold overdigestion with BclI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH. 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA, and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Blocked dcm methylation: Not sensitive CpG methylation: Not sensitive
|
Reagents supplied: 10x M and 10x K buffer |
||||||||||||||||||||||||||||||||||||||
BamHI
| BamH I |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
BamHI is a restriction enzyme purified from Bacillus amyloliquefaciens H.
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 100 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 5 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 100 units of BamHI incubated for 16 hours at 37oC with 1 μg of λ DNA resulted in a DNA pattern free of detectable nuclease degradation as determined by agarose gel electrophoresis. After 50-fold overdigestion with BamHI, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 80oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
Star activity: Conditions of low ionic strength, high enzyme concentration, glycerol concentration>5% or pH>8.0 may result in star activity. |
Reagents supplied: 10x UBamHI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
AsuII (isoschizomer)
| Asu II (isoschizomer) |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
Asu II is a restriction enzyme purified from an isolated strain (#94S).
Unit substrate: Lambda DNA (Hind III digest).
Unit calculation assay conditions: 50 mM NaCl, 10 mM Tris-HCl (pH 7.9 @ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100, 100 μg/ml BSA. Incubate at 37oC.
Absence of contaminants: 100 units of AsuII do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA/Hind III digest at 37oC. After 50-fold overdigestion with AsuII, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.9@ 25°C), 0.1 mM EDTA, 1 mM dithiothreitol, 0.15% Triton X-100, 200 μg /ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked
|
Reagents supplied: 10x UAsuII and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||
ApaLI
| ApaL I |  Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
ApaL I is a restriction enzyme purified from Acetobacter pasteurianus (ATCC 12875).
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 100 units of ApaL I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of lambda DNA at 37oC. After 100-fold overdigestion with ApaL I, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol. Store at -20oC.
Heat inactivation: No.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive
|
Reagents supplied: 10x L and 10x K buffer |
||||||||||||||||||||||||||||||||||||||
pUC19 DNA/Hpa II Digest
|
pUC19 DNA/Hpa II Digest |
 Technical Brochure Technical Brochure |
| Description: The Hpa II (BsiS I) digest of pUC19 DNA yields the following 13 discrete fragments (in base pairs): 501, 489, 404, 331, 242, 190, 147, 111, 110, 67, 34, 34, 26.
Preparation: pUC19 DNA was completely digested by BsiS I, phenol/chloroform extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
 |
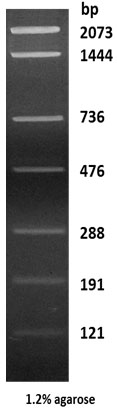
pUC19 DNA/BseBI/TaqI Digest
|
pUC19 DNA/BseBI/TaqI Digest |
 Technical Brochure Technical Brochure |
| Description: This digest of pUC19 DNA yields the following 9 discrete fragments (in base pairs): 2073, 1444, 736, 476, 288, 191, 121, 30, 13.
Preparation: pUC19 DNA was completely digested by BseBI, pUC19 DNA was completely digested by TaqI, mixed, phenol/chloroform extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
 |
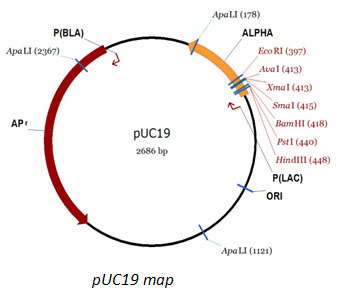
pUC19 DNA
|
pUC19 DNA |
 Technical Brochure Technical Brochure |
| Description: pUC19 is a commonly used plasmid cloning vector in E. coli. The molecule is double-stranded circle, 2686 base pairs in length, and has a high copy number. pUC19 carries a 54 base-pair multiple cloning site polylinker that contains unique sites for 13 different hexanucleotide-specific restriction endonucleases. The molecular weight of pUC19 is 1.75x106 daltons.
Preparation: pUC19 is isolated from E. coli strain HB101 by a standard plasmid purification procedure.
Quality control: Gel analysis for purity. BsiSI and AluI fragmentation patterns.
Storage buffer: 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
 |
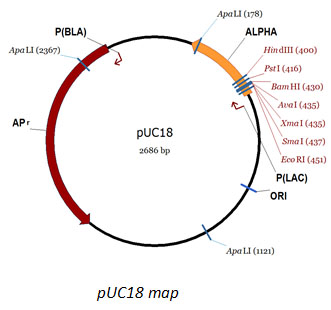
pUC18 DNA
|
pUC18 DNA |
 Technical Brochure Technical Brochure |
|
Description: pUC18 is a commonly used plasmid cloning vector in E. coli. The molecule is double-stranded circle, 2686 base pairs in length, and has a high copy number. pUC18 carries a 54 base-pair multiple cloning site polylinker that contains unique sites for 13 different hexanucleotide-specific restriction endonucleases. The molecular weight of pUC18 is 1.75x106 daltons.
Preparation: pUC18 is isolated from E. coli strain HB101 by a standard plasmid purification procedure.
Quality control: Gel analysis for purity. BsiSI and AluI fragmentation patterns.
Storage buffer: 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
 |
pBR322 DNA HinfI Digest
|
pBR322 DNA HinfI Digest |
 Technical Brochure Technical Brochure |
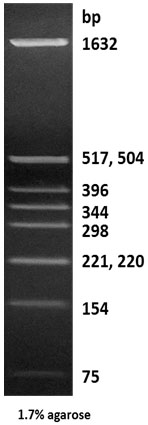
|
Description: The Hinf I digest of pBR322 DNA yields the following 10 discrete fragments (in base pairs): 1632, 517, 504, 396, 344, 298, 221, 220, 154, 75.
Preparation: pBR322 DNA was completely digested by Hinf I, phenol/chloroform extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
 |
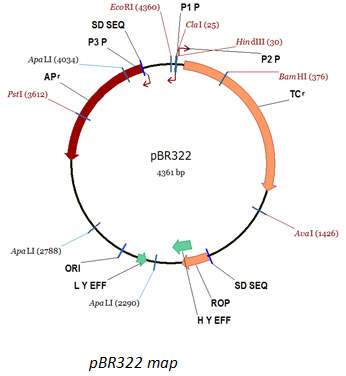
pBR322 DNA
|
pBR322 DNA |
 Technical Brochure Technical Brochure |
| Description: pBR322 is a commonly used plasmid cloning vector in E. coli. The molecule is double-stranded circle, 4361 base pairs in length. pBR322 contains the genes for resistance to ampicillin and tetracycline, and can be amplified with chloramphenicol. The molecular weight of pBR322 is 2.83x106 daltons.
Preparation: pBR322 is isolated from E. coli strain HB101 by a standard plasmid purification procedure.
Quality control: Gel analysis for purity. HinfI and AluI fragmentation patterns.
Storage buffer: 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
 |
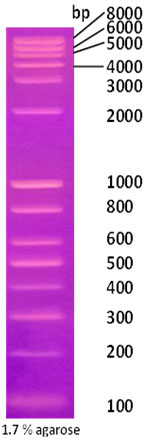
Long Range DNA Ladder marker
|
Long Range DNA Ladder marker |
 Technical Brochure Technical Brochure |
| Description: The Long Range DNA Ladder is suitable for sizing double-stranded DNA from 100bp to 8kb. The 1kb band has increased intensity to serve as reference point. The double-stranded ladder can be visualized on 1% to 2% agarose gels after ethidium bromide staining.
Preparation: Complete digestion of MINOTECH biotechnology plasmid mix with appropriate restriction enzyme yields the following 14 discrete fragments (in kilobase pairs): 8, 6, 5, 4, 3, 2, 1, 0.8, 0.6, 0.5, 0.4, 0.3, 0.2, 0.1.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
Related products: 10X Loading Dye Solution |
 |
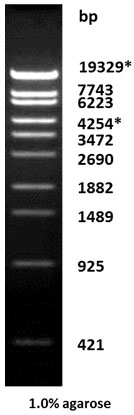
Lambda DNA StyI Digest
|
Lambda DNA StyI Digest |
 Technical Brochure Technical Brochure |
| Description: The Sty I digest of λ DNA yields the following 11 discrete fragments (in base pairs): 19329*, 7743, 6223, 4254*, 3472, 2690, 1882, 1489, 925, 421, 74.
Preparation: Lambda DNA was completely digested by Sty I, phenol extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
*The cohesive ends (12b cos site of bacteriophage λ) of fragments 19329bp and 4254bp may anneal to and form the additional band. These fragments may be separated by heating to 65oC for 5 min and then cooling on ice for 3min. |
|
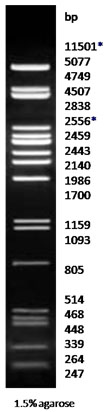
Lambda DNA PstI Digest
|
Lambda DNA PstI Digest |
 Technical Brochure Technical Brochure |
|
Description: The Pst I digest of λ DNA yields the following discrete fragments (in base pairs): 11501*, 5077, 4749, 4507, 2838, 2556*, 2459, 2443, 2140, 1986, 1700, 1159 1093, 805, 514, 468, 448, 339, 264, 247, 216, 211, 200, 164, 150, 94, 87, 72, 15.
Preparation: Lambda DNA was completely digested by Pst I, phenol extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
*The cohesive ends (12b cos site of bacteriophage λ) of fragments 11501bp and 2556bp may anneal to and form the additional band. These fragments may be separated by heating to 65oC for 5 min and then cooling on ice for 3min.
Related products: 10X Loading Dye Solution
|
 |
Lambda DNA Hind III Digest
|
Lambda DNA Hind III Digest |
 Technical Brochure Technical Brochure |
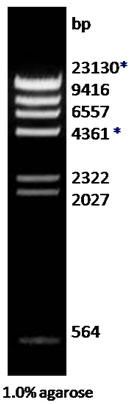
| Description: The Hind III digest of λ DNA yields the following 8 discrete fragments (in base pairs): 23130*, 9416, 6557, 4361*, 2322, 2027, 564, 125.
Preparation: Lambda DNA was completely digested by Hind III, phenol extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated..
*The cohesive ends (12b cos site of bacteriophage λ) of fragments 23130 and 4361 may be separated by heating to 65oC for 5 minutes.
Related products: 10X Loading Dye Solution |
 |
Lambda DNA/EcoR I Digest
|
Lambda DNA/EcoR I Digest |
 Technical Brochure Technical Brochure |
|
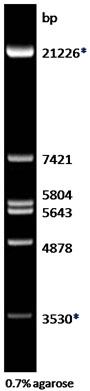
Description: The EcoR I digest of λ DNA yields the following 6 discrete fragments (in base pairs): 21226*, 7421, 5804, 5643, 4878, 3530*.
Preparation: Lambda DNA was completely digested by EcoR I, phenol extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
*The cohesive ends (12b cos site of bacteriophage λ) of fragments 21226bp and 3530bp may anneal to and form the additional band. These fragments may be separated by heating to 65oC for 5 min and then cooling on ice for 3min.
Related products: 10X Loading Dye Solution
|
 |
Taq DNA Polymerase
|
Taq DNA Polymerase |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||||||||||
|
Source: Purified from an E. coli strain carrying a plasmid with Taq DNA polymerase gene from Thermus aquaticus YT-1.
Description: Taq DNA Polymerase is a thermostable enzyme that catalyzes 5'→3' synthesis of DNA. The enzyme has no detectable 3'→5' proofreading exonuclease activity, but possesses low 5'→3' exonuclease activity. Unit definition: One unit is defined as the amount of enzyme required to catalyze the incorporation of 10 nmoles of dNTPs into acid insoluble material in 30 minutes at 72oC.
Reaction conditions: 1x Taq polymerase buffer [50 mM KCl, 10 mM Tris-HCl pH 8.5 @ 25oC, 1.5 mM MgCl2, 0.1% Triton X-100].
Quality Control Assays:
|
Guaranteed stability: Taq DNA polymerase is guaranteed to maintain stability for six months from the date of shipment when stored as directed. Storage Buffer: 100 mM NaCl, 50 mM Tris-HCl (pH 8.0 @ 25oC), 1 mM DTT, 0.1 mM EDTA, 1% Triton X-100 and 50% glycerol. Store at -20oC.
Reagents supplied: 10x Taq DNA polymerase Buffer (1.5ml) and/or 10x Taq DNA polymerase w/o MgCl2 (1.5ml) and 25mM MgCl2 (1.5ml).
|
||||||||||||||||||||||||||||||||||||||||||||||
T4 DNA Ligase
|
T4 DNA Ligase *Add an H to cat.# to order the high concentration |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| Source: T4 DNA ligase is purified from E. coli lambda lysogen NM 989.
Description: T4 DNA Ligase catalyzes the formation of a phosphodiester bond between juxtaposed 5′-phosphate and 3′-hydroxyl termini in duplex DNA or RNA.
Unit definition: One Weiss unit is defined as the amount of enzyme required to catalyze the exchange of 1 nmol of 32P from pyrophosphate to ATP, into Norit-adsorbable material in 20 minutes at 37°C
Reaction conditions: 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP (not included) and DNA (recommended DNA concentration 0.1 to 1 μM of 5´ termini). Οptimal ligation occurs at 16oC.
Quality control: Tested for the absence of endo- and exodeoxyribonucleases, ribonucleases and for the capacity to join cohesive- and blunt-ended DNA fragments.
Storage buffer: 50 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: T4 DNA Ligase can be inactivated by incubation at 65oC for 10 minutes.
Notes
|
Recommended ligation mixtures:
Reagents supplied: 10x Ligase Reaction buffer (w/o ATP) |
||||||||||||||||||||||||||||||||||||||
DNA Methyltransferase M.BseCI
|
DNA Methyltransferase M.BseCI |
 Technical Brochure Technical Brochure |
|
5'…ATCGAT…3'
Description: M.ΒseCI modifies the N6 atom of the 3΄ adenine residue in the sequence 5΄-ATCGAT-3΄.
Source: M.BseCI is a methyltransferase purified from an E. coli strain that carries the BseCI methyltransferase gene (bseCIM) from Bacillus stearothermophilus, cloned in plasmid pBseCIM8 (1,2).
Reaction Buffer: 10 mM Tris-HCl (pH 7.4), 10 mM EDTA, 5 mM 2-mercaptoethanol, 0.02% Triton-X-100. Unit definition: One unit is defined as the amount of enzyme required to protect 1μg of λ DNA in 1 hour at 55oC in a total reaction volume of 10μl against cleavage by BseCI restriction endonuclease. Protection Assay Conditions: M.BseCI is incubated with 1μg of λ DNA in 10μl 1x M.BseCI buffer, supplemented with 80μM S-adenosylmethionine (SAM), for 1 hour at 55oC followed by 15 minutes at 70oC. The extent of protection by M.BseCI is determined by the addition of 40μl BseCI Reaction Buffer and 10 units of BseCI restriction endonuclease. Incubation at 55oC for 30 minutes is followed by analysis on agarose gel. Note: M.BseCI exhibits 35% activity at 37oC. Storage buffer: 50 mM Tris-HCl (pH 7.4), 10 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC. |
Quality Control: Tested for the absence of endo- and exodeoxyribonucleases.
References: 1. Rina, M. and Bouriotis, V. (1993) Cloning, purification and characterization of the BseCI DNA methyltransferase from Bacillus stearothermophilus. Gene 133, 91-94. 2. Rina, M., Markaki, M. and Bouriotis, V. (1994) Sequence of the cloned bseCIM gene: M.BseCI reveals high homology to M.BanIII Gene 150, 71-73.
Reagents supplied: 10x M.BseCI buffer |
Lambda DNA EcoR I/Hind III Digest
|
Lambda DNA EcoRI/HindIII Digest |
 Technical Brochure Technical Brochure |
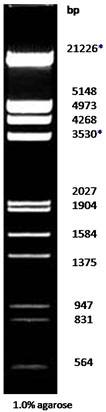
| Description: The EcoR I and Hind III digest of λ DNA yields the following 13 discrete fragments (in base pairs): 21226*, 5148, 4973, 4268, 3530*, 2027, 1904, 1584, 1375, 947, 831, 564, 125.
Preparation: Lambda DNA was completely digested by EcoR I and Hind III, phenol extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
*The cohesive ends (12b cos site of bacteriophage λ) of fragments 21226bp and 3530bp may anneal to and form the additional band. These fragments may be separated by heating to 65oC for 5 min and then cooling on ice for 3min.
Related products: 10X Loading Dye Solution |
 |
Lambda DNA BstE II Digest
|
Lambda DNA BstE II Digest |
 Technical Brochure Technical Brochure |
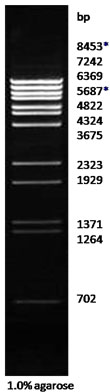
| Description: The BstE II digest of λ DNA yields the following 14 discrete fragments (in base pairs):
8453*, 7242, 6369, 5687*, 4822, 4324, 3675, 2323, 1929, 1371, 1264, 702, 224, 117.
Preparation: Lambda DNA was completely digested by BstE II, phenol extracted, ethanol precipitated, dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
*The cohesive ends (12b cos site of bacteriophage λ) of fragments 8453bp and 5687bp may anneal to and form an additional band of 14140bp. These fragments may be separated by heating to 65oC for 5 min and then cooling on ice for 3min.
Related products: 10X Loading Dye Solution |
 |
Lambda DNA
| Lambda DNA (concentration 0.5μg/μl) |
|
|
Preparation: The phage is isolated from the heat inducible lysogen E. coli λ cl857 S7 by gel filtration. The DNA is isolated from the purified phage by phenol/chloroform extraction and dialyzed against 10 mM Tris-HCl (pH 7.4) and 1 mM EDTA.
Quality control: Gel analysis for purity. EcoRI and HindIII fragmentation patterns.
Storage buffer: 10 mM Tris-HCl (pH 7.4), 1 mM EDTA. Store at -20oC.
Related products: 10X Loading Dye Solution |
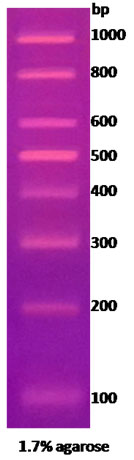
100bp DNA Ladder marker
|
100bp DNA Ladder marker |
 Technical Brochure Technical Brochure |
| Description: The 100 bp DNA Ladder is suitable for sizing double-stranded DNA from 100 to 1000 bp. The 500 and 1,000 base pair bands have increased intensity to serve as reference points. The double-stranded ladder can be visualized on 1% to 2% agarose gels after ethidium bromide staining.
Preparation: Complete digestion of MINOTECH biotechnology 100bp ladder plasmid with appropriate restriction enzyme yields the following 8 discrete fragments (in base pairs): 1000, 800, 600, 500, 400, 300, 200, 100.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
Related products: 10X Loading Dye Solution |
 |
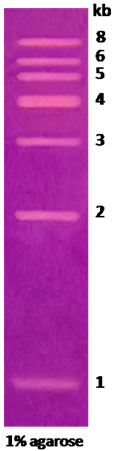
1kb DNA Ladder marker
|
1kb DNA Ladder marker |
 Technical Brochure Technical Brochure |
| Description: The 1kb DNA Ladder is suitable for sizing double-stranded DNA from 1kb to 8kb. The 4kb band has increased intensity to serve as reference point. The double-stranded ladder can be visualized on 1% to 2% agarose gels after ethidium bromide staining.
Preparation: Complete digestion of MINOTECH biotechnology plasmid mix with appropriate restriction enzyme yields the following 7 discrete fragments (in kilobase pairs): 8, 6, 5, 4, 3, 2, 1.
Storage buffer: 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Store at -20oC.
Note: Dilute in TE or other buffer of minimal ionic strength. DNA may denature if diluted in dH2O and subsequently heated.
Related products: 10X Loading Dye Solution |
 |
DNA Polymerase I Large Fragment (Klenow Fragment)
| DNA Polymerase I Large Fragment (Klenow Fragment) |
 Technical Brochure Technical Brochure |
Description: The Klenow Fragment lacks the 5′→3′ exonuclease activity of intact DNA Polymerase I but retains the 5′→3′ polymerase, the 3′→5′ exonuclease and the strand displacement activities. Source: Purified from an E. coli strain carrying a DNA Polymerase I large fragment overproducing plasmid. 1X Klenow Reaction Buffer: Reaction conditions: 50 mM Tris-HCl (pH 7.6 @ 25oC), 5 mM MgCl2, 1 mM DTT and dNTPs. Klenow fragment is also 50% active in all five standard MINOTECH buffers when supplemented with dNTPs. Unit definition: One unit is defined as the amount of enzyme required to convert 10 nmoles of dNTPs to an acid insoluble form in 30 minutes at 37oC. Quality control: The enzyme is greater than 98% pure as indicated by SDS-polyacrylamide gel electrophoresis and contains no detected endonuclease activity. Incubation of 10U of Klenow with supercoiled plasmid DNA produced no nicked molecules after 20 hours at 37oC as determined by agarose gel electrophoresis analysis. |
Storage buffer: 0.1 M KPO4 (pH 6.5), 1 mM DTT and 50% glycerol. Store at -20oC.
Heat inactivation: 75oC for 20 minutes. Fill-in conditions: Dissolve 0.1-4 μg of digested DNA in 1x Klenow reaction buffer supplemented with 40 μM each dNTP. Add 1 unit Klenow per μg DNA and incubate 15 minutes at 25oC. Stop the reaction by adding EDTA to 10 mM final concentration and heating at 75oC for 10 minutes. Note: excessive amounts of enzyme or longer reaction times may result in recessed ends due to the 3′→5′ exonuclease activity of the enzyme. Reagents supplied: 10x Klenow Reaction buffer |
AluI
| AluI |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
|
AluI is a restriction enzyme purified from Arthrobacter luteus (ATCC 21606).
Unit substrate: Lambda DNA.
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.9 @ 25oC), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin and DNA. Incubate at 37oC.
Absence of contaminants: 50 units of AluI do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of lambda DNA at 37oC. After 10-fold overdigestion with Alu I, greater than 95% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 100 mM KCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml bovine serum albumin and 50% glycerol.Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Not sensitive
|
Reagents supplied: 10x L and 10x K buffer |
||||||||||||||||||||||||||||||||||||||