|
SgrBI (SacII isoschizomer) |
 Technical Brochure Technical Brochure |
||||||||||||||||||||||||||||||||||||||
| SgrBI is a restriction enzyme purified from Streptomyces griseus.
Unit substrate: Lambda DNA (HindIII digest).
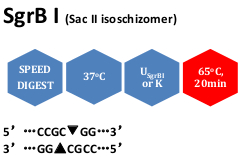
Unit calculation assay conditions: 10 mM Tris-HCl (pH 7.9@ 25°C), 10 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100, 100 μg/ml BSA. Incubate at 37°C.
Absence of contaminants: 400 units of SgrB I do not produce any unspecific cleavage products after 16 hrs incubation with 1 μg of λ DNA (HindIII digest) at 37°C. After 100-fold overdigestion with SgrBI, greater than 98% of the DNA fragments can be ligated and recut with this enzyme.
Storage buffer: 50 mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg/ml BSA and 50% glycerol. Store at -20oC.
Heat inactivation: 65oC for 20 minutes.
Methylation Sensitivity: dam methylation: Not sensitive dcm methylation: Not sensitive CpG methylation: Blocked
Note: Particular sites in λ and φΧ174 DNAs are difficult to cleave with SgrB I, as well as with its prototype Sac II. Reference: Rina, M., Pagomenou, M. and V, Bouriotis (1991) Nucleic Acids Res. 19, 6342. |
Reagents supplied: 10x USgrBI and 10x K buffer
|
||||||||||||||||||||||||||||||||||||||

